Professional Documents
Culture Documents
Ex No.-03 Polymerase Chain Reaction
Uploaded by
Sujit ShandilyaOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Ex No.-03 Polymerase Chain Reaction
Uploaded by
Sujit ShandilyaCopyright:
Available Formats
Ex No.
- 03 Polymerase Chain Reaction
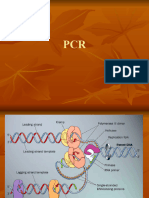
Aim:To amplify the isolated DNA by PCR. Principles:PCR allows amplification of the target DNA into millions of copies by following repetitive cycling of 3 specific successive steps, each time doubling the DNA molecule. The final number of copies of the DNA can be expressed by the formula. 2nx m Where, n=no. of cycle m=no. of copies of original template PCR amplification involves three major steps which is done in an automated thermal cycler which can heat and cool the tubes with the reaction mixture in a very short time. The temperature and time can be set accordingly to the requirement of each reaction. Steps in PCR are explained as below. 1. Denaturation at 94C:During this step, the double strand melts open to single stranded DNA , all enzymatic reaction stop. 2. Annealing at 54C:During this step Oligonucleotide primer anneals with the single stranded DNA Oligonucleotide constantly try to attach with the complementary bases in the dissociated template DNA by hydrogen bond. The more stable bonds last a little bit longer (primer that fit exactly) and on that little piece of DNA (template & primer), the polymerase can attach and start copying the
template. Once there are a few bases built in the hydrogen bond is so strong between the template and the primers do not break anymore. 3. Extension at 72C:This is the ideal working temperature for the polymerase, where the primers extend in the 3direction with polymerase in the presence of dNTPs. Primers that are on positions with no exact match get loose again (because of the higher temperature) and dont give extension of the fragment. The bases (complementary to the template) are coupled to the primer on the 3side (the polymerase adds dNTPs from 5 to 3 reading the template from 3 to 5side, bases are added complementary to the template. Materials: Template DNA (genomic, plasmid, cosmid, bacterial/yeast colony, etc.) Primers (resuspended to a known concentration with sterile TE) Buffer (usually 10X, usually sold with Taq polymerase or you can make your own) Note: some different buffer receipes follow at the end of this protocol MgCl2 (25mM is convenient) Taq polymerase dNTPs (2mM stock) Note: a 2mM stock of dNTPs means that the final concentration of each dNTP (dATP, dCTP, dGTP, and dTTP) is 2mM -- NOT that all dNTPs together make 2mM. dNTPs come as 100mM stocks -- thaw and add 10L of each dNTP to 460L of ddH20 to make 2mM. Store at -20C.
Reaction Components of PCR:1. DNA polymerase: Most commonly used DNA polymerase is Taq polymerase which is isolated from a bacterium found in hot spring known as Thermus aquaticus. It works optimally at 72C and over pH range 7.0-7.5, adding 100 nucleotides per second to the primer. It is a heat stable enzyme withstanding repeated Denaturation cycles.
2. Deoxynucleotide triphosphates (dNTPs): It is available as a mixture of deoxyadinosine triphosphates (dATP), deoxycytidine triphosphates (dCTP), deoxyguanosine triphosphates (dGTP) and deoxythymidine triphosphates (dTTP). Usually each dNTP is used at concentration between 50m and 200m. these dNTPs attach to the free 3 hydroxyl group of the primer and form a complementary to the template strand. 3. Reaction buffer and MgCl2 The buffer most often used in PCR is 10mM Tris buffer with pH range of 8.5-9.0 at 25C. Because the pH of the Tris buffer decreases by 0.3 units by each 10C rise in the temperature, a buffer made to pH 8.8 at 25C will have a pH of 7.4 at 72C. The dNTPs bind Mg2+ and hence the reaction mixture must contain excess Mg2+. Generally the Mg2+ concentration in the reaction mixture is 0.5 to 2.5mM greater than the concentration of dNTPs. Concentration of Mg2+ also influences the efficiency of primer to template annealing. 4. PCR Primers: Primer concentration should not be greater than 1m. In general, primers should be 20-30 nucleotides in length, which allows reasonably high annealing temperature. The primer should be made with an approximately equal number of each of the 4 bases avoiding regions of unusual sequence such as polypurines, polypyrimidines or repetitive motifs. There should be no 3 end complementarily between primer pairs to avoid primer-dimer formation. 5. Template DNA: Intact template DNA is a prerequisite of successful PCR. Over abundance of template DNA will favor annealing of the 2 strands of template sequence rather than annealing to primer pair and thereby reduce the efficiency of PCR. PCR Protocol:The following reagents were added to the PCR tubes to a final volume of 50l. Use 2l of 60ng/l or 100ng/l DNA
Use 2l of each primer at 3.2pmole/l concentration or 2.5l of each primer at 100ng/l concentration 5l 10x PCR Buffer w/ Mg 1l 25mM MgCl2 1l dNTP 0.25l Taq 36.75l sterile water to equal a 50l rxn (*if not making a master mix, dilute Taq so that you can add 1l of Taq and 36l sterile water to equal a 50l rxn) 1. Keep the reagents on ice. 2. Add the Taq last, and keep it in the freezer until you are ready to add it. 3. Vortex briefly and quick spin. 4. Cycle: 95C for 1-5minutes (usually 4min) 95C for 1min 55C for 1min 72C for 1.5 to 2min (usually 2min) 72C for 10min 4C hold Cycle 30 times
You might also like
- Temperature-Responsive Polymers: Chemistry, Properties, and ApplicationsFrom EverandTemperature-Responsive Polymers: Chemistry, Properties, and ApplicationsNo ratings yet
- TYPES OF PCR - Dr. De-1.pdfDocument33 pagesTYPES OF PCR - Dr. De-1.pdfJAICHANDRUNo ratings yet
- Gel Electrophoresis of ProteinsFrom EverandGel Electrophoresis of ProteinsMichael J DunnNo ratings yet
- PCR ManualDocument6 pagesPCR ManualjiaNo ratings yet
- PCR RTPCR, QPCRDocument88 pagesPCR RTPCR, QPCRshravani sahuNo ratings yet
- Principle of The PCR: This Technique Is Invented in 1984 by Kary Mulis. The Purpose of A PCRDocument11 pagesPrinciple of The PCR: This Technique Is Invented in 1984 by Kary Mulis. The Purpose of A PCRantesar reheemNo ratings yet
- Polyme Rase Chain Reactio N: Course Code: 501Document23 pagesPolyme Rase Chain Reactio N: Course Code: 501Humayun ArshadNo ratings yet
- MKBS221. PRAC 5 REPORT Last OneDocument11 pagesMKBS221. PRAC 5 REPORT Last OneNOMKHULEKO ALICENo ratings yet
- PCRDocument34 pagesPCRSumitNo ratings yet
- Molecular Diagnostics Techniques Polymerase Chain ReactionDocument7 pagesMolecular Diagnostics Techniques Polymerase Chain ReactionJamesNo ratings yet
- PcrDocument35 pagesPcrk5zo7jNo ratings yet
- PCR LectureDocument18 pagesPCR LecturePagla HowaNo ratings yet
- Polymerase Chain Reaction: Description of ComponentsDocument5 pagesPolymerase Chain Reaction: Description of ComponentsSamirSmsmNo ratings yet
- The Polymerase Chain Reaction (PCR) : BackgroundDocument4 pagesThe Polymerase Chain Reaction (PCR) : BackgroundМ.К. МариNo ratings yet
- Gene-specific RT-PCR optimizationDocument4 pagesGene-specific RT-PCR optimizationQuimico Inmunologia GeneticaNo ratings yet
- Lab Manual 3Document7 pagesLab Manual 3syazaismailNo ratings yet
- PCRDocument42 pagesPCRNopiyana PujiastutiNo ratings yet
- Analiza PCR - ThermocyclerDocument7 pagesAnaliza PCR - Thermocyclerionescu_ion_3No ratings yet
- Polymerase Chain Reaction (PCR)Document37 pagesPolymerase Chain Reaction (PCR)Haleema SultanNo ratings yet
- The Polymerase Chain Reaction (PCR)Document50 pagesThe Polymerase Chain Reaction (PCR)Deana NamirembeNo ratings yet
- 03 PCRDocument5 pages03 PCRToni D.No ratings yet
- Polymerase Chain Reaction: Description of ComponentsDocument5 pagesPolymerase Chain Reaction: Description of ComponentsSkand BhardwajNo ratings yet
- BB2111 PCR TOBEPRINTEDDocument7 pagesBB2111 PCR TOBEPRINTEDapi-3839553No ratings yet
- PCR Overview GoldBioDocument1 pagePCR Overview GoldBioKIARAH EUNIZE GICANo ratings yet
- PCR Amplification of DNA SequencesDocument56 pagesPCR Amplification of DNA SequencesSamiksha SharmaNo ratings yet
- Inverse PCRDocument6 pagesInverse PCRHiromi UchimaNo ratings yet
- Polymerase Chain ReactionDocument19 pagesPolymerase Chain ReactionKamila Haidari100% (1)
- Aula TBM5 2022-2023 - EnglishDocument29 pagesAula TBM5 2022-2023 - EnglishDiogo Botelho Medeiros BrilhanteNo ratings yet
- Polymerase Chain Reaction (PCR)Document21 pagesPolymerase Chain Reaction (PCR)greateinsteinNo ratings yet
- Bio Project-Polymerase Chain ReactionDocument21 pagesBio Project-Polymerase Chain ReactionS.AbiniveshNo ratings yet
- Polymera SE Chain ReactionDocument6 pagesPolymera SE Chain ReactionpriyaNo ratings yet
- Morgan 1922: 2000 Genes in 4 Chromosome of D.: MelanogasterDocument44 pagesMorgan 1922: 2000 Genes in 4 Chromosome of D.: MelanogasterlordniklausNo ratings yet
- PCR Lecture HURO Zador Final SmallDocument59 pagesPCR Lecture HURO Zador Final SmallvsripathirajaNo ratings yet
- Module 4 - PCRDocument12 pagesModule 4 - PCRnavneetNo ratings yet
- General PCR ProtocolDocument2 pagesGeneral PCR ProtocolSanju KaramNo ratings yet
- Polymerase Chain ReactionDocument47 pagesPolymerase Chain ReactionshvnagaNo ratings yet
- 2X Taq FroggaMix PCR KitDocument4 pages2X Taq FroggaMix PCR KitCésar CuadraNo ratings yet
- PCR Labwork 2 ENGDocument4 pagesPCR Labwork 2 ENGmigas1996No ratings yet
- 2× Direct Blood PCR Master Mix: Catalog: Sample Pack Store at - 20°C Blood StorageDocument2 pages2× Direct Blood PCR Master Mix: Catalog: Sample Pack Store at - 20°C Blood StoragepazhanivelNo ratings yet
- Prerequisite of A PCR: Logarithmic and Controlled FashionDocument18 pagesPrerequisite of A PCR: Logarithmic and Controlled FashionlkokodkodNo ratings yet
- Viva 2 StepsDocument4 pagesViva 2 StepsElimar CarreonNo ratings yet
- Polymerase Chain Reaction (PCR)Document3 pagesPolymerase Chain Reaction (PCR)biotech_vidhyaNo ratings yet
- DNA amplification 2020Document5 pagesDNA amplification 2020Blameless ArikoNo ratings yet
- Dream Taq Green MM #k1081Document2 pagesDream Taq Green MM #k1081victor_david_19No ratings yet
- The Olymerase Hain Eaction: Zhang-Haifeng Department of BiochemistryDocument42 pagesThe Olymerase Hain Eaction: Zhang-Haifeng Department of Biochemistryapi-19916399No ratings yet
- Polymerase Chain ReactionDocument5 pagesPolymerase Chain ReactionAaryan DeswalNo ratings yet
- Radio Immuno As SayDocument6 pagesRadio Immuno As SayhardmanpersonNo ratings yet
- Polymerase Chain ReactionDocument27 pagesPolymerase Chain ReactionundalliNo ratings yet
- Chaudhary Charan Singh University Meerut (U.P.) : "PCR & Microarray"Document33 pagesChaudhary Charan Singh University Meerut (U.P.) : "PCR & Microarray"desma elitaNo ratings yet
- Introduction PCR AssignmentDocument20 pagesIntroduction PCR AssignmentogunfunminiyiestherNo ratings yet
- Polymerase Chain ReactionDocument13 pagesPolymerase Chain Reactionahmad ghNo ratings yet
- Polymerase Chain ReactionDocument13 pagesPolymerase Chain Reactionvamshibommi3477No ratings yet
- Nucleic Acid TestingDocument55 pagesNucleic Acid TestingShaiji ShahidNo ratings yet
- mecA gene allows antibiotic resistanceDocument5 pagesmecA gene allows antibiotic resistanceANNAALTSHULERNo ratings yet
- Components of PCRDocument3 pagesComponents of PCRpmc9q544vwNo ratings yet
- Standard PCR Protocol: How To Do PCRDocument7 pagesStandard PCR Protocol: How To Do PCRCarla FernandesNo ratings yet
- Brief Notes On Polymerase Chain Reaction (PCR) : 2 Year MT Molecular Biology Lab 2010Document5 pagesBrief Notes On Polymerase Chain Reaction (PCR) : 2 Year MT Molecular Biology Lab 2010kinantiNo ratings yet
- NUCLEIC ACID AMPLIFICATIONDocument6 pagesNUCLEIC ACID AMPLIFICATIONbsramos2023No ratings yet
- PCR An OverviewDocument41 pagesPCR An OverviewnassradabourNo ratings yet
- New Microsoft Office Word DocumentDocument1 pageNew Microsoft Office Word DocumentSujit ShandilyaNo ratings yet
- Glossary of CommunicationDocument7 pagesGlossary of CommunicationSujit ShandilyaNo ratings yet
- Sugar Fermentation TestDocument10 pagesSugar Fermentation TestSujit ShandilyaNo ratings yet
- Assignment PrimerDocument5 pagesAssignment PrimerSujit ShandilyaNo ratings yet
- Sujit The Lactic Acid BacteriaDocument4 pagesSujit The Lactic Acid BacteriasushanfcriNo ratings yet
- Impact FactorDocument1 pageImpact FactorSujit ShandilyaNo ratings yet
- Science Vs ReligionDocument1 pageScience Vs ReligionSujit ShandilyaNo ratings yet
- HACCP Principles and Applications: Introduction To Food Safety HazardsDocument6 pagesHACCP Principles and Applications: Introduction To Food Safety HazardsSujit ShandilyaNo ratings yet
- Littopenaeus Vannamei SujitDocument16 pagesLittopenaeus Vannamei SujitSujit ShandilyaNo ratings yet
- HACCPDocument47 pagesHACCPSujit Shandilya100% (1)
- Cell BiologyDocument3 pagesCell BiologySujit ShandilyaNo ratings yet
- How To Calculate Freezing PointDocument3 pagesHow To Calculate Freezing PointSujit ShandilyaNo ratings yet
- How To Calculate Freezing PointDocument3 pagesHow To Calculate Freezing PointSujit ShandilyaNo ratings yet
- Bio EnergeticsDocument7 pagesBio EnergeticsSujit ShandilyaNo ratings yet
- Dna Ligase, Terminal Transferase, Adapter and LinkerDocument80 pagesDna Ligase, Terminal Transferase, Adapter and LinkerSujit ShandilyaNo ratings yet
- Assignmrnt of Fish Nutrition SujitDocument10 pagesAssignmrnt of Fish Nutrition SujitSujit ShandilyaNo ratings yet
- Biodiversity ConservationDocument4 pagesBiodiversity ConservationSujit ShandilyaNo ratings yet
- TopoisomeraseDocument7 pagesTopoisomeraseSujit ShandilyaNo ratings yet
- Bycatch Reduction Devices - PresentationDocument92 pagesBycatch Reduction Devices - PresentationSujit Shandilya0% (1)
- Weekly Schedule TemplateDocument1 pageWeekly Schedule TemplateSujit ShandilyaNo ratings yet
- Waste Recycling and Fish CultureDocument19 pagesWaste Recycling and Fish CultureSujit Shandilya100% (1)
- Bio EnergeticsDocument7 pagesBio EnergeticsSujit ShandilyaNo ratings yet
- Classification of MarketsDocument2 pagesClassification of MarketsSujit Shandilya100% (3)
- Nutrient-Gene Interactions in FishesDocument5 pagesNutrient-Gene Interactions in Fishesrahulraj1122No ratings yet
- Fish Biotech Courses 2nd Sem MFScDocument1 pageFish Biotech Courses 2nd Sem MFScSujit ShandilyaNo ratings yet
- Kola FisheryDocument1 pageKola FisherySujit ShandilyaNo ratings yet
- Top 100 journals by impact factorDocument332 pagesTop 100 journals by impact factorJaisson Miyosi OkaNo ratings yet
- Cell BiologyDocument3 pagesCell BiologySujit ShandilyaNo ratings yet
- Indian Fishery Genetics - History in Breaf Sujit Kumar, Cife MumbaiDocument15 pagesIndian Fishery Genetics - History in Breaf Sujit Kumar, Cife MumbaiSujit ShandilyaNo ratings yet
- Central Dogma - STR of Dna and Its Different Form, Transcription, TranslationDocument16 pagesCentral Dogma - STR of Dna and Its Different Form, Transcription, TranslationSujit ShandilyaNo ratings yet
- Degenerate PrimerDocument2 pagesDegenerate PrimerAmod KumarNo ratings yet
- Determining The Diet of Larvae of Western Rock LobDocument11 pagesDetermining The Diet of Larvae of Western Rock LobCherie QuintoNo ratings yet
- BTG 402 - BIOTECHNOLOGY - BHU2022-2023 - NotesDocument47 pagesBTG 402 - BIOTECHNOLOGY - BHU2022-2023 - NotesEmmanuella OffiongNo ratings yet
- Important MCQ Colloction For PartDocument154 pagesImportant MCQ Colloction For PartNazia Ali86% (7)
- Science10 Q3 Week4Document22 pagesScience10 Q3 Week4Samad Recca T. NatividadNo ratings yet
- Gallaeformans Ditylenchus Sp.Document18 pagesGallaeformans Ditylenchus Sp.Edgar Medina GomezNo ratings yet
- Module 3 Activity Central DogmaDocument5 pagesModule 3 Activity Central DogmaNORODIN DALANDAS0% (1)
- Biotechnology8 q3 Mod1 Tools-in-Genetic-EngineeringDocument20 pagesBiotechnology8 q3 Mod1 Tools-in-Genetic-Engineeringunc bNo ratings yet
- QPCR Primer Design RevisitedDocument10 pagesQPCR Primer Design RevisitedkuangNo ratings yet
- Abi P Bigdye Primer Cycle Sequencing Ready Reaction Kit: ProtocolDocument43 pagesAbi P Bigdye Primer Cycle Sequencing Ready Reaction Kit: ProtocolhadymatrixNo ratings yet
- Addgene: Protocol - How To Design PrimersDocument1 pageAddgene: Protocol - How To Design PrimersArantxa Ortega LeonNo ratings yet
- Target Amplification MethodsDocument3 pagesTarget Amplification MethodsNOR-FATIMAH BARATNo ratings yet
- The Molecular Basis of InheritanceDocument35 pagesThe Molecular Basis of Inheritanceprehealthhelp80% (5)
- Identification and Phylogeny of Ascomycetous Yeasts From Analysis of Nuclear Large Subunit (26S) Ribosomal DNA Partial SequencesDocument41 pagesIdentification and Phylogeny of Ascomycetous Yeasts From Analysis of Nuclear Large Subunit (26S) Ribosomal DNA Partial SequencesJokanoe LertNo ratings yet
- SundariDocument8 pagesSundariRani Eva DewiNo ratings yet
- 4-6 Seegene Bulletin Vol1 SDocument17 pages4-6 Seegene Bulletin Vol1 SjoseNo ratings yet
- KOD - Plus-: Store at - 20°CDocument7 pagesKOD - Plus-: Store at - 20°CMNo ratings yet
- 2009-Comparative Sensitivity of PCR Primer Sets For Detection of Cryptosporidium ParvumDocument6 pages2009-Comparative Sensitivity of PCR Primer Sets For Detection of Cryptosporidium ParvumWiwien HendrawanNo ratings yet
- Chromosome AberrationDocument37 pagesChromosome AberrationAli Abbas AslamNo ratings yet
- Plant Rarity Lab ReportDocument15 pagesPlant Rarity Lab Reportapi-529961637No ratings yet
- Test Bank For Essential Cell Biology 4th Edition Bruce AlbertsDocument32 pagesTest Bank For Essential Cell Biology 4th Edition Bruce Albertsharoldbrownorcnxeijgd100% (26)
- Summary Notes - Topic 6 Microbiology, Immunity and Forensics - Edexcel (IAL) Biology A-LevelDocument11 pagesSummary Notes - Topic 6 Microbiology, Immunity and Forensics - Edexcel (IAL) Biology A-LevelSaeed AbdulhadiNo ratings yet
- Site Directed MutagenesisDocument77 pagesSite Directed MutagenesisDanny Sebastian Thomas100% (1)
- New York Sars-Cov-2 Real-Time Reverse Transcriptase (RT) - PCR Diagnostic PanelDocument27 pagesNew York Sars-Cov-2 Real-Time Reverse Transcriptase (RT) - PCR Diagnostic PanelcassNo ratings yet
- Description: Sensifast™ Cdna Synthesis KitDocument2 pagesDescription: Sensifast™ Cdna Synthesis KitDevin HendrawanNo ratings yet
- Acanthamoeba Strains Isolated From Organs of Freshwater FishesDocument8 pagesAcanthamoeba Strains Isolated From Organs of Freshwater FishestishaNo ratings yet
- Central Dogma of Molecular BiologyDocument30 pagesCentral Dogma of Molecular BiologyAlthea Mandal100% (1)
- Basic Principles and Components of PCRDocument30 pagesBasic Principles and Components of PCRDiahNo ratings yet
- Buckingham - Molecular Diagnostics-Fundamentals Methods and Clinical ApplicationsDocument479 pagesBuckingham - Molecular Diagnostics-Fundamentals Methods and Clinical Applicationsfakefacebook75891% (11)
- 7.1 - Nucleic Acids, Ahl: (Adapted From)Document5 pages7.1 - Nucleic Acids, Ahl: (Adapted From)Big CinemaNo ratings yet
- 10% Human: How Your Body's Microbes Hold the Key to Health and HappinessFrom Everand10% Human: How Your Body's Microbes Hold the Key to Health and HappinessRating: 4 out of 5 stars4/5 (33)
- When the Body Says No by Gabor Maté: Key Takeaways, Summary & AnalysisFrom EverandWhen the Body Says No by Gabor Maté: Key Takeaways, Summary & AnalysisRating: 3.5 out of 5 stars3.5/5 (2)
- Why We Die: The New Science of Aging and the Quest for ImmortalityFrom EverandWhy We Die: The New Science of Aging and the Quest for ImmortalityRating: 3.5 out of 5 stars3.5/5 (2)
- Crypt: Life, Death and Disease in the Middle Ages and BeyondFrom EverandCrypt: Life, Death and Disease in the Middle Ages and BeyondRating: 4 out of 5 stars4/5 (3)
- This Is Your Brain On Parasites: How Tiny Creatures Manipulate Our Behavior and Shape SocietyFrom EverandThis Is Your Brain On Parasites: How Tiny Creatures Manipulate Our Behavior and Shape SocietyRating: 3.5 out of 5 stars3.5/5 (31)
- The Ancestor's Tale: A Pilgrimage to the Dawn of EvolutionFrom EverandThe Ancestor's Tale: A Pilgrimage to the Dawn of EvolutionRating: 4 out of 5 stars4/5 (811)
- The Consciousness Instinct: Unraveling the Mystery of How the Brain Makes the MindFrom EverandThe Consciousness Instinct: Unraveling the Mystery of How the Brain Makes the MindRating: 4.5 out of 5 stars4.5/5 (93)
- The Other Side of Normal: How Biology Is Providing the Clues to Unlock the Secrets of Normal and Abnormal BehaviorFrom EverandThe Other Side of Normal: How Biology Is Providing the Clues to Unlock the Secrets of Normal and Abnormal BehaviorNo ratings yet
- All That Remains: A Renowned Forensic Scientist on Death, Mortality, and Solving CrimesFrom EverandAll That Remains: A Renowned Forensic Scientist on Death, Mortality, and Solving CrimesRating: 4.5 out of 5 stars4.5/5 (397)
- The Lives of Bees: The Untold Story of the Honey Bee in the WildFrom EverandThe Lives of Bees: The Untold Story of the Honey Bee in the WildRating: 4.5 out of 5 stars4.5/5 (44)
- Undeniable: How Biology Confirms Our Intuition That Life Is DesignedFrom EverandUndeniable: How Biology Confirms Our Intuition That Life Is DesignedRating: 4 out of 5 stars4/5 (11)
- The Molecule of More: How a Single Chemical in Your Brain Drives Love, Sex, and Creativity--and Will Determine the Fate of the Human RaceFrom EverandThe Molecule of More: How a Single Chemical in Your Brain Drives Love, Sex, and Creativity--and Will Determine the Fate of the Human RaceRating: 4.5 out of 5 stars4.5/5 (515)
- Human Errors: A Panorama of Our Glitches, from Pointless Bones to Broken GenesFrom EverandHuman Errors: A Panorama of Our Glitches, from Pointless Bones to Broken GenesRating: 3.5 out of 5 stars3.5/5 (56)
- Wayfinding: The Science and Mystery of How Humans Navigate the WorldFrom EverandWayfinding: The Science and Mystery of How Humans Navigate the WorldRating: 4.5 out of 5 stars4.5/5 (18)
- The Mind & The Brain: Neuroplasticity and the Power of Mental ForceFrom EverandThe Mind & The Brain: Neuroplasticity and the Power of Mental ForceNo ratings yet
- A Brief History of Intelligence: Evolution, AI, and the Five Breakthroughs That Made Our BrainsFrom EverandA Brief History of Intelligence: Evolution, AI, and the Five Breakthroughs That Made Our BrainsRating: 4.5 out of 5 stars4.5/5 (4)
- Eels: An Exploration, from New Zealand to the Sargasso, of the World's Most Mysterious FishFrom EverandEels: An Exploration, from New Zealand to the Sargasso, of the World's Most Mysterious FishRating: 4 out of 5 stars4/5 (30)
- Gathering Moss: A Natural and Cultural History of MossesFrom EverandGathering Moss: A Natural and Cultural History of MossesRating: 4.5 out of 5 stars4.5/5 (347)
- The Second Brain: A Groundbreaking New Understanding of Nervous Disorders of the Stomach and IntestineFrom EverandThe Second Brain: A Groundbreaking New Understanding of Nervous Disorders of the Stomach and IntestineRating: 4 out of 5 stars4/5 (17)
- Darwin's Dangerous Idea: Evolution and the Meaning of LifeFrom EverandDarwin's Dangerous Idea: Evolution and the Meaning of LifeRating: 4 out of 5 stars4/5 (523)
- Younger for Life: Feel Great and Look Your Best with the New Science of AutojuvenationFrom EverandYounger for Life: Feel Great and Look Your Best with the New Science of AutojuvenationRating: 4 out of 5 stars4/5 (1)
- Inside of a Dog: What Dogs See, Smell, and KnowFrom EverandInside of a Dog: What Dogs See, Smell, and KnowRating: 4 out of 5 stars4/5 (390)
- Superlative: The Biology of ExtremesFrom EverandSuperlative: The Biology of ExtremesRating: 4.5 out of 5 stars4.5/5 (51)
- The Magic of Reality: How We Know What's Really TrueFrom EverandThe Magic of Reality: How We Know What's Really TrueRating: 4.5 out of 5 stars4.5/5 (106)
- Lymph & Longevity: The Untapped Secret to HealthFrom EverandLymph & Longevity: The Untapped Secret to HealthRating: 4.5 out of 5 stars4.5/5 (13)