Professional Documents
Culture Documents
Study Guide T2 BioChem2
Uploaded by
janeCopyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Study Guide T2 BioChem2
Uploaded by
janeCopyright:
Available Formats
Study Guide Test 2
Q: Strategies of nucleotide synthesis:
Strategies of nucleotide synthesis. There are two general strategies for nucleotide
synthesis: de novo (from precursors) or salvage pathways from bases (without ribose or
phosphates). These pathways are often targets of anticancer drugs.
Q: Precursors, when is ribose added, how is ribose activated and added, what are first purines
and pyrimidines made
PURINES
IMP is the first purine synthesized, and other purines are synthesized from IMP.
The strategy of purine nucleotide synthesis is to start with ribose and build the purine
on the ribose atom by atom.
Activation of the ribose is the first step. Ribose-5-phosphate pyrophosphokinase
catalyzes pyrophosphate addition to the C-1 of ribose to form 5-phosphoribosyl-pyrophosphate (PRPP), an important intermediate in de novo purine synthesis, de novo
pyrimidine synthesis, and all the salvage pathways. Because this is an important
intermediate in several pathways, it is not the first committed step in purine synthesis.
The first committed step is the formation of phosphoribosyl-amine from PRPP with
glutamine as a nitrogen donor.
Both purines are synthesized from IMP in two steps.
AMP synthesis requires replacement of an oxygen with a nitrogen, with aspartate as
the nitrogen donor. The addition of aspartate requires GTP. The same enzyme removes
the four-carbon unit in synthesis of IMP and AMP.
GMP synthesis involves addition of an extra nitrogen. This first involves addition of an
oxygen, followed by an ATP-dependent replacement of the oxygen with a nitrogen
with glutamine as the nitrogen donor.
PYRIMIDINES
Strategy. The strategy is the reverse of that for purine synthesis. The pyrimidine ring is
synthesized before addition of the ribose. Remember for purines, the ribose provides a
scaffold for purine synthesis.
1
Synthesis. Only two molecules contribute to the pyrimidine ring: carbamoyl phosphate,
and aspartate
Q: How are the various forms of THF made and/or interconverted
There are six forms of C1-THF, which exist at different oxidation states, and are
interconvertible by NADP(H) oxidation or reductions.
Most is generated by conversion of serine to glycine.
Q: Regulation of purine synthesis (where are committed steps), how is a balance between AMP
and GMP achieved, how do folic acid antagonists impair purine synthesis.
Some of the regulation has been discussed and is summarized in Fig. 26.6. PRPP
also activates the second step, and such regulation is sometimes called feedforward.
In addition to these early steps, there is regulation at the IMP branch. AMP
inhibits the first step in IMP to AMP, and GMP inhibits the first step in IMP to
GMP. Also notice that GTP is a substrate for AMP synthesis and ATP is a
substrate for GMP synthesis. The net effect of these levels of regulation is to
maintain a proper balance of AMP and GMP.
Formation of nucleoside di- and triphosphates, adenylylate (AMP) kinase and ATP generation in
muscle.
Q: Purine salvage pathways: the role of PRPP, and the two major phosphoribosyltransferases,
The nucleobases, adenine, guanine, and hypoxanthine (the base for inosine), are normal
products of metabolism. This is described in the next section. It is energetically better to
make nucleotides from the bases than from the de novo pathways. This is done with two
phosphoribosyltransfersases, which add a phosphoribosyl group from PRPP to the
bases to form AMP, GMP, and IMP.
The reaction is base + PRPP NMP + PPi.
There are two phosphoribosyl transferases: adenine (APRT) and hypoxanthine-guanine
(HGPRT).
Q: Lesch-Nyhan disease, gout and uric acid.
2
A complete deficiency of HGPRT results in a disease called Lesch-Nyhan syndrome.
The disease causes uric acid accumulation, which results in gouty arthritis. (Uric acid is a
purine degradation product, which forms when purines are not salvaged.) The disease
results in severe defects in the nervous system, and produces mental retardation,
aggressive behavior, and self-mutilation.
The gene is on the X chromosome, and only occurs in males.
The failure to salvage purines causes an enormous increase in purine synthesis, which
may result from elevated PRPP.
The disease is easily detected in fetal cells following amniocentesis.
Q: Nucleic acid degradation in intestine (fate of the base), and in cells; first common
intermediate in degradation of all purines, the two reactions of xanthine oxidase, the use of
allopurinol.
SCID, adenosine deaminase deficiency, and resulting metabolic problems.
The function of the purine nucleoside cycle.
Pyrimidine synthesis: precursors, first pyrimidine made, first cytosine derivative made,
regulation, metabolic channeling
Deoxyribonucleotide synthesis and ribonucleotide reductase: source of reducing power,
thioredoxin, substrates, binding of substrates to catalytic site, the two regulatory sites.
Synthesis of dTMP: precursor, the THF derivatives (substrate and products), and the unusual
reaction of a THF derivative, why is this enzyme a target of inhibitors of DNA synthesis and how
do folic acid analogs block synthesis.
Q: Tautomerizations. How do they affect base pairing, how do they cause mutations?
Nucleotide base can occur as either of two alternative tautonomers, structural isomers
that are in dynamic equilibrium. The shift in structure determines whether Nitrogen or
Oxygen is hydrogen-bond acceptors or donors. The Oxygen and Nitrogen determine base
pairing properties.
Tautomeric-based structural changes are frequent causes of mutations. If the rare
tautonomer base pairs incorrectly, A:C and G:T base pairs can occur.
Q: What bonds in nucleotides and nucleic acids are unstable and under what conditions?
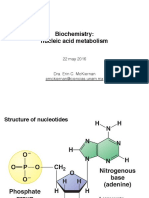
Q: Know the difference between bases, nucleosides, nucleotides, and their names.
Names of nucleosides are derived by adding osine to the purine bases, and an idine to
the pyrimidines.
Bases are: Adenine, Guanine, Cytosine, Uracil and Thymine.
Nucleosides are the bases + the ribose
Nucleotides have a phosphate esterified to a hydroxyl group of the ribose. For the
common nucleotide, esterification is at 5OH
Q: The glycosidic linkage and its stability.
Nucleosides are bases linked to a sugar via a glycosidic linkage.
Anomeric Carbon is attached to the N-1 of the pyrimidine and N-9 of the purine.
It is in the beta configuration (above the ring) for all nucleosides and nucleotides.
The glycosidic linkage in nucleosides is stable in alkali solutions. However in acid
condition, the purine glycosidic linkage is labile, but pyridine linkage is stable.
Q: Anti and Syn configurations: what structure is not allowed and why?
Syn configuration has the base over the sugar; Anti conformation is fully extended and
does not have base over sugar.
Pyrimidines favor anti configuration because their Oxygen gets in the way.
Purines can adopt either syn or anti configuration.
Q: The biological function of adenosine, effects of caffeine.
Generally nucleosides do not accumulate, and have no distinctive biological function
The only exception is Adenosine, which can act as a local hormone (autocoid).
4
Adenosine circulates in the bloodstream and influences blood vessel dilatation, smooth
contraction, neural discharge, neurotransmitter release, and fat metabolism.
Hard-working muscle cells release adenosine, which dilates blood vessels, increase blood
flow, O2 delivery and nutrient delivery.
Regulates heart beats by blocking the electrical current that controls the heart, which
slows heart beats.
Also involved in sleep regulation. After long periods of wakefulness, brain adenosine
levels rise which promote sleepiness. Caffeine counteracts this effect by blocking the
interaction of adenosine with the neural receptors.
Conventions for writing polynucleotides sequences and structures.
Q: Classes of nucleic acids and their properties. What are unique structures or components of
each type of RNA?
DNA
Double helix with strands running in opposite orientation, with base pairs held togerher
with hydrogen bonds.
It lacks the 2OH
RNA
Three basic types with rRNA being the most abundant one.
Also other forms such as snRNA in eukaryotes and tmRNA in prokaryotes.
mRNA
Made from one strand of DNA
In prokaryotes, a single strand mRNA may contain the information for several proteins.
This is rarely the case for eukaryotic mRNA.
Eukaryotic mRNA is transcribed as hnRNA first and then processed to smaller forms
(splicing).
Introns are removes and exons remain to be translated.
Eukaryotic mRNA have a run of 100-200 As, called polyA tail, which adds stability. They
are added after transcription.
Bacterial mRNAs also have As, but these actually promote degradation.
5
rRNA
Most abundant RNA
About 65% of weight of Ribosomes.
Contain number of unusual bases.
tRNAs
Carriers of amino acids for protein synthesis.
They are highly folded due to intramolecular Hydrogen bonding
Many of they bases are modified.
Amino acid is attached at the 3end.
Some amino acids may have up to five different amino acids.
There are different tRNAs in each compartment (cytoplasm versus mitochondria).
snRNA
Are small nuclear RNAs.
Many are modified
Abundant in nucleus of eukaryotes
Assoiciated with proteins and participate in splicing.
Small RNAs
They can bind to RNA or DNA by complementarity
Can be used for gene silencing.
Q: Nucleic acid stability in acid and alkaline solution.
In highly acidic solution (pH<2.3) bases are protonated, which disrupts Hydrogen
bonding.
In highly alkaline solution (pH>10) the bases are deprotonated and hydrogen bonding is
disrupted.
Nucleic acid hydrolysis and nuclease specificity: where does cleavage occur?
Hydrolysis often breaks the phosphodiester backbones.
Acid Hydrolysis: 1mM HCl hydrolyzes the purine glycosidic linkage in DNA but not
RNA. The phosphodiester bond is intact but a gap exists in the DNA.
Alkaline hydrolysis: DNA is stable but RNA is not. Phosphate may end up in 2 or 3
carbon of ribose.
Nucleases is a hydrolysis enzyme
Nucleases degrade nucleic acids. These enzymes are found in all cells. Snake venom
contains a potent phosphodiesterase, which breaks phosphodiester backbones. Cleavage
can occur on either side of phosphate.
A convention labels the sites of cleavage as a for the 3 side, and b for the 5 side.
Nucleases are also categorized as endo or exo according to where they work on.
Specificity of Nucleases
Some nucleases attach on ly DNA or RNA. Some attack both
Can be specific towards single or double stranded nucleic acids.
Others require binding to a specific sequence, such as restriction enzymes.
Restriction enzymes and specific endonucleases.
Q: The logic of nucleic acid sequencing by the Sanger method and 454 technology.
Dideoxy method, Sangers method:
This method depends on synthesis of DNA.
Primer is added. It is complementary to a specific sequence on a single stranded template.
Primer must have a free 3-OH which permits chain elongation.
DNA polymerases add nucleosides. Here an altered form of DNA Polymerase is used. It
lacks exanuclease activity (proof reading function).
All four deoxynucleotides are added in four reaction tubes. One of these nucleotides is
targeted. In each tube, a different 2, 3 dideoxynucleotide is added.
There is little of dideoxynucleotide in the solution, but when it is incorporated, the chain
synthesis stops because it lacks 3OH.
This generates aseries of oligonucleotides with different lengths, which are separated by
size with gel lectrophoresis. The small frgsmenst travel further and fastest.
454 Technology
Primer plus DNA is immobilized on beads. Beads are loaded into very small wells
DNA Polymerase plus one nucleotide, wash then
DNA plus a second nucleotide, wash then third and fourth in separate steps
Up to 500 cycles of the four steps.
7
When a nucleotide is added, light is emitted. Phosphate is released, and the first reaction
of sulfate reduction is reversed to form ATP and sulfate.
ATP plusluciferin emits light.
Q: The structural differences between A-, B-, and Z-DNA. Do these occur inside cells?
A and B DNA are right-handed helices; Z DNA is a right-handed helix.
A-DNA
The pitch is 2.46 nm and is 11 bp.
Not naturally occurring. However, dehydrated DNA and DNA:RNA hybrids may assume
this conformation.
B-DNA
A:T and G:C base pairs have virtually the same size.
Base pair spacing is 0.34 nm
The helical repeat 9one pitch) is 10 to 10.6 base pairs long and 3.4 nm.
Hydrogen bonding is possible if strands are antiparallel
Polar sugar backbones are outside.
The hydrophobic bases are on the outside.
Helical twisting brings base pairs closer.
In Z-DNA;
C residues assume syn rather than anti conformation. To accommodate the change, C
residues must flip, and it takes the sugar with it. In the next bp, the purine is the anti
conformation.
The sugar phosphate backbone zigzags.
Methylation of C allows Z-DNA formation even if purines and pyridines do not alternate.
Q: Alternative hydrogen-bonded structures: how can cruciforms form; hoogsteen base pairs and
triplexes; what are the factors that favor quadruplexes and where are they found?
Cruciforms: an inverted repeat on a single strand can base pait and form cruciform. These
structures may form in cells, and may be sites for the binding of proteins.
Hoogsteen base pairs. There are different ways of hydrogen-bonding. Normally, it
involves interactions between two six-membered rings. However, h-bonding can also
involve the five-membered rings. This alternate bonding creates a hoogsteen base pair.
DNA quadruplex structures are G-rich, cyclic and have hoogsteen base interaction.
Certain cations (k+, Na+, Ca2+) favor these structures via the O6 carbonyl group. Variety
of these structures can form. The are involved in Telomeres, Gene rearrangements, gene
regulatory regions, and some human diseases.
Q: Factors that affect denaturation and renaturation.
DNA denaturation
A variety of conditions can destabilize hydrogen bonds and result in strand separation:
changes in pH, temperature, and ionic strength. The strands are said to be denatured.
If temperature separates strands, the strands are said to have melted.
As the strands denature, the absorbance at 260 nm increases by up to 40%. This is called
the hyperchromic shift. When the bases are stacked, the potential to absorb UV light is
reduced since the pi electrons in the aromatic rings are constrained. Unstacking removes
the constraints. The midpoint in the absorbance increase is called the melting temperature
(or Tm).
The Tm is a function of G:C content: higher Tm with higher G:C content.
Increasing ionic strength increases stability. This results from decreasing the electrical
repulsion of the phosphates. DNA readily denatures in low ionic strength solutions.
At pH > 10, the bases are deprotonated, and H-bonding is disrupted.
Similarly, at pH < 2.3, the bases are protonated, which disrupts H-bonding.
Small solutes that readily form H bonds also denature DNA. Examples include urea and
formamide.
Renaturation
Denatured DNA will renature if the disrupting agent is removed.
The strand must reassociate in a process called reannealing.
Once the proper register is found (this is slow), the process can be very rapid.
The process is facilitated by a moderate temperature (just below melting), which permits
rapid testing of different associations, until the right register is found.
The rate depends on the complexity of the DNA. It takes longer for larger DNA to find
the right register than short DNA. More complex DNA takes longer to associate.
Nucleic acid hybridizations and DNA complexity.
DNA tertiary structure: supercoils, topoisomerases, and gyrases
Nucleosomes and higher order structures of DNA.
Structures of tRNA and rRNA: major features.
Features of DNA replication: semiconservative, bidirectional, role of helicases and
topoisomerases, semidiscontinuous synthesis, Okazaki fragments
Properties of DNA polymerases: the various activities, core and holoenzymes, the -complex and
its function, the clamp loader and the sliding clamp, the differences between DNP I and III and
their functions.
Q: Telomere replication
The sequence form protective caps (1-12 kbp) at the end of chromosomes.
They consist of short (5-8 bp) G-rich sequences that are repeated.
Vertebrate telomeres are TTAGG.
DNP cannot make the extreme 5 end because a primer is needed.
Lagging strand synthesis at the 3ends of chromosomes is primed by RNA primase,
leading to Okazaki fragment synthesis. The primer is removed, and a gap is created.
Telomerase is an RNA-dependent DNA Polymerase that restores this missing sequence.
The enzyme contains RNA that serves as a template.
In humans, this component is 450 nucleotides long.
The 3 end of the DNA is used as a primer to add the repeats.
What are the causes of errors during DNA replication?
10
Bases is in the incorrect tautonomer, and therefore mispair. In these mispairings, an Hbond acceptor can tautomerize to an H-bond donor. Proofread function of DNPIII usually
catches these erros.
A in syn base pairs with G.
T:C base pairs mediated by water.
Mutations can be also induced by mutagens. For example, a base analog can be
incorporated into DNA, and be designated to cause mispairing.
Q: How can you induce mutations?
Mutations can be induced by mutagens.
5-Bromouracil (5-BU) is Thymine analog. It tautomerizes frequently, and base pairs
frequently with G instead of A, which induces a transition.
2-aminopurine (2-AP) acts similarly. It occasionalu base pairs with C instead of T,
causing a transition.
Hypoxantine can arise in situ by oxidative deamination of adenine. Hypozanthine base
pairs with C, causing transition.
Mutagens can be base modifying agents.
Nitrogenous acid (HNO2) deaminates A to C. C deamination creates Uracil, which
changes a C:G to a U:A and eventually to a T:A pair.
Hydeoxylamine reacts with C, converting it to a derivative that base pairs with A.
Alkylating agents add methyl or ethyl groups to bases, which alter their H-bonding
properties. O6-methyl-G pairs with T. These agents can also cause transversions. They
can labilize the N-glycosidic linkage of G, which leaves a gap in the DNA. A repair
enzyme, AP endonuclease, cleaves the phosphodiester backbone, the DNA on one strand
is removed, and a new nucleotide is inserted. A transversion results if a pyrimidine is
inserted.
DNA repair mechanisms.
Be able to distinguish:
Nitrogenous bases:
Adenine
Guanine
Hypoxanthine
11
Cytosine
Uracil
Thymine
Nucleoside:
Adenosine
Guanosine
Inosine
Uridine
Cytidine
Thymidine
Nucleotides:
adenylic acid (AMP), GMP, IMP, UMP, CMP
dAMP, dGMP, dIMP, dUMP, dGMP, dTMP
12
You might also like
- Synthesis and Degra NuclotidesDocument58 pagesSynthesis and Degra Nuclotidesur.yared21No ratings yet
- Amino Acid Protein and Nucleic Acid Metabolism 20182019 LectureDocument18 pagesAmino Acid Protein and Nucleic Acid Metabolism 20182019 LectureMwanja MosesNo ratings yet
- Purine Metabolism PDFDocument29 pagesPurine Metabolism PDFtrinitysugumar0% (1)
- DR Okunowo Wahab Introductory Molecular Biology Lecture Note I (Nucleotides Metabolism)Document20 pagesDR Okunowo Wahab Introductory Molecular Biology Lecture Note I (Nucleotides Metabolism)modelprof100% (2)
- Metabolism of Nucleoproteins Part IDocument50 pagesMetabolism of Nucleoproteins Part IAgafioNo ratings yet
- Nucleotide Metabolism PPDocument65 pagesNucleotide Metabolism PPCLEMENTNo ratings yet
- BSC Sem VI Nuc Aid MetabDocument24 pagesBSC Sem VI Nuc Aid Metabstarboigaming08No ratings yet
- Metabolic Hemostasis MF PortionDocument68 pagesMetabolic Hemostasis MF PortionTsegaye HailuNo ratings yet
- PG Purine and PyrimidineDocument14 pagesPG Purine and PyrimidineAbdulnafiu AhmadNo ratings yet
- Metabolism of NucleotidesDocument24 pagesMetabolism of NucleotidesAditya NayakNo ratings yet
- Met Purin 28 OktDocument62 pagesMet Purin 28 OktXIID 67No ratings yet
- Nucleic Acid MetabolismDocument23 pagesNucleic Acid MetabolismMSc Biotech/MicroNo ratings yet
- NucleotideDocument56 pagesNucleotideDhara NPNo ratings yet
- Disorders of Purine and Pyrimidine MetabolismDocument17 pagesDisorders of Purine and Pyrimidine Metabolismtanmay mehtaNo ratings yet
- Presentacion EntenderDocument37 pagesPresentacion EntenderJuan David Marin ChiguachiNo ratings yet
- Nukleotida: Dr. I Dewa Ayu Susilawati, Drg. M. KesDocument33 pagesNukleotida: Dr. I Dewa Ayu Susilawati, Drg. M. KesAldiansyahHakimNo ratings yet
- Nucleic AcidsDocument48 pagesNucleic AcidsMelissa SalayogNo ratings yet
- HJHJJJJJDocument91 pagesHJHJJJJJShrey SundriyalNo ratings yet
- Purine SynthesisDocument51 pagesPurine Synthesisdr.yogita.rajNo ratings yet
- NUS 123 NotesDocument14 pagesNUS 123 NotesBrevine Oduor OsoroNo ratings yet
- Metabolism of NucleoproteinsDocument71 pagesMetabolism of NucleoproteinsMi PatelNo ratings yet
- Nukleotida: Dr. I Dewa Ayu Susilawati, Drg. M. KesDocument33 pagesNukleotida: Dr. I Dewa Ayu Susilawati, Drg. M. KesIkatanti Ratna AnggrainiNo ratings yet
- Chapter 18 - Nucleotide Metabolism: Nucleotides Are Composed of Three ComponentsDocument5 pagesChapter 18 - Nucleotide Metabolism: Nucleotides Are Composed of Three ComponentsSaiful IslamNo ratings yet
- Drug Metabolism - Chapter 8Document44 pagesDrug Metabolism - Chapter 8Shaun李好No ratings yet
- Pyrimidine SynthesisDocument35 pagesPyrimidine Synthesisdr.yogita.rajNo ratings yet
- L 3 Proteins Peptide Bond FormationDocument21 pagesL 3 Proteins Peptide Bond FormationAhmed Zubair IrshadNo ratings yet
- Trans of Harper's Illustrated Biochemistry CH 32Document3 pagesTrans of Harper's Illustrated Biochemistry CH 32Richelle Dianne Ramos-Giang100% (1)
- Biochemistry - Metabolism of Purines and PyrimidinesDocument11 pagesBiochemistry - Metabolism of Purines and PyrimidinesProjjal SanyalNo ratings yet
- 12 เมแทบอลิซึมของกรดนิวคลีอิกDocument23 pages12 เมแทบอลิซึมของกรดนิวคลีอิก22vk6svnb4No ratings yet
- ACAWPurineand Pyrimindine Synthesis Presentationfor October 112010Document41 pagesACAWPurineand Pyrimindine Synthesis Presentationfor October 112010Ezekoko ChineseNo ratings yet
- 3study Questions-Drug MetabolismDocument5 pages3study Questions-Drug Metabolismapi-3723612No ratings yet
- VCE Biology Summary Book 2013: Samantha TierneyDocument102 pagesVCE Biology Summary Book 2013: Samantha Tierneymickyhearts18No ratings yet
- Purine Metabolism de Novo Synthesis and Salvage Pathway, 2015Document28 pagesPurine Metabolism de Novo Synthesis and Salvage Pathway, 2015Tehreem NadeemNo ratings yet
- BAGIAN PERTAMA - Metabolisme Asam NukleatDocument84 pagesBAGIAN PERTAMA - Metabolisme Asam NukleatDendy FyransyahNo ratings yet
- Week VII - Nucleotide Metabolism - Pptx-Merged-CompressedDocument332 pagesWeek VII - Nucleotide Metabolism - Pptx-Merged-Compressedjessenia.tutor2010No ratings yet
- Ytochrome Nzymes: Presented by Deshmukh MD Faizan M. Pharm (1 Sem)Document20 pagesYtochrome Nzymes: Presented by Deshmukh MD Faizan M. Pharm (1 Sem)Anitha Mary DambaleNo ratings yet
- Biosintesis Asam Amino & Nukleotida: Puji Lestari Prodi Kimia FST UNSOEDDocument35 pagesBiosintesis Asam Amino & Nukleotida: Puji Lestari Prodi Kimia FST UNSOEDJennie JisooNo ratings yet
- Nucleotide Metabolism: By: Rebecca Asis Villanueva M.D. Associate Professor Department of Biochemistry & NutritionDocument82 pagesNucleotide Metabolism: By: Rebecca Asis Villanueva M.D. Associate Professor Department of Biochemistry & NutritionproiskNo ratings yet
- Biochemistry - Nucelic AcidsDocument96 pagesBiochemistry - Nucelic AcidsSandra OnceNo ratings yet
- CSIR-NET Life Science - Short Notes On Nucleotide Biosynthesis (Purine & Pyrimidine Synthesis)Document10 pagesCSIR-NET Life Science - Short Notes On Nucleotide Biosynthesis (Purine & Pyrimidine Synthesis)Arun DubeyNo ratings yet
- Biochemistry Chapter 5Document10 pagesBiochemistry Chapter 5Jayson AguilarNo ratings yet
- Nucleic Acid MetabolismDocument35 pagesNucleic Acid MetabolismAmreen KhanNo ratings yet
- Curs Nucleotide Sem 2Document58 pagesCurs Nucleotide Sem 2George PetreaNo ratings yet
- Biomedic I-Biochemistry: Department of Biochemistry. Faculty of Medicine Universitas HasanuddinDocument72 pagesBiomedic I-Biochemistry: Department of Biochemistry. Faculty of Medicine Universitas HasanuddinZafa AiharaNo ratings yet
- Biochemistry Chapter 5Document9 pagesBiochemistry Chapter 5Jayson AguilarNo ratings yet
- BP U9e Metabolism of Nucleic AcidsDocument49 pagesBP U9e Metabolism of Nucleic AcidsChristian Angelo AgbunagNo ratings yet
- Biochem Metabolismo Ácidos NucléicosDocument27 pagesBiochem Metabolismo Ácidos NucléicosLidia Escutia GuadarramaNo ratings yet
- 3.unit IDocument53 pages3.unit IHarshiniNo ratings yet
- Purine Biosynthesis and RegulationDocument25 pagesPurine Biosynthesis and RegulationrivienaNo ratings yet
- Biochemistry ReviewDocument7 pagesBiochemistry ReviewMinh MinhNo ratings yet
- MicrobiologyDocument18 pagesMicrobiologymbkal3liNo ratings yet
- Metabolism of Purine and PyrimidineDocument56 pagesMetabolism of Purine and PyrimidineAboubakar Moalim Mahad moh'dNo ratings yet
- Chemistry of Nucleotides and Nucleic AcidsDocument72 pagesChemistry of Nucleotides and Nucleic AcidsBernard100% (1)
- Study Guide Test 2 Biochem UTDDocument2 pagesStudy Guide Test 2 Biochem UTDJenny JiouNo ratings yet
- Nucleotide Biosynthesis Lecture 2017sDocument23 pagesNucleotide Biosynthesis Lecture 2017sSaad KazmiNo ratings yet
- Chapter 3 - MBDocument31 pagesChapter 3 - MBMustee TeferaNo ratings yet
- Purine MBBSDocument28 pagesPurine MBBSDikpal BikramNo ratings yet
- Nucleic AcidDocument88 pagesNucleic AcidJoyce Jimenez100% (1)
- Lec.6 PPP Pathway PDFDocument5 pagesLec.6 PPP Pathway PDFhawraaNo ratings yet
- Preclinical Biochemistry and Medical Genetics Review 2023: For USMLE Step 1 and COMLEX-USA Level 1From EverandPreclinical Biochemistry and Medical Genetics Review 2023: For USMLE Step 1 and COMLEX-USA Level 1No ratings yet
- ,, ,, - 3, - Omega-Q AriixDocument3 pages,, ,, - 3, - Omega-Q AriixOlga VoicuNo ratings yet
- Reactions of Alkenes and AlkynesDocument2 pagesReactions of Alkenes and AlkynesanthonyNo ratings yet
- Experiment 9 - Vitamin Analysis PDFDocument14 pagesExperiment 9 - Vitamin Analysis PDFAALIYAH REIGN MAPUSAONo ratings yet
- Applications of ChemistryDocument23 pagesApplications of ChemistryLearn With UsNo ratings yet
- Amino Acid CatabolismDocument31 pagesAmino Acid CatabolismLidya YudithNo ratings yet
- Antoine ConstantsDocument1 pageAntoine Constantsradwaelhadad75% (8)
- Using My Fitness PalDocument3 pagesUsing My Fitness Palpep pepeNo ratings yet
- Kode Barang Nama Barang Stok HargaDocument10 pagesKode Barang Nama Barang Stok HargayemimaNo ratings yet
- Nutri PrelimsDocument15 pagesNutri PrelimsGabNo ratings yet
- New Dyslipidemia 2021 Naplex QuickDocument2 pagesNew Dyslipidemia 2021 Naplex Quickkaylakmills_1013586883% (6)
- HACCP Carbonated Soft DrinksDocument9 pagesHACCP Carbonated Soft DrinksmubdiantiNo ratings yet
- 4A Alkenes and Alkanes TestDocument5 pages4A Alkenes and Alkanes TestMinorNo ratings yet
- Stock PPG Tangerang Mahakam, Kalbe & Hexpharm 07 Nov 23Document22 pagesStock PPG Tangerang Mahakam, Kalbe & Hexpharm 07 Nov 23rian agustianNo ratings yet
- Soaps and SyndetbarsDocument18 pagesSoaps and Syndetbars329 Vishal Sharma civilNo ratings yet
- لیست کد های ATCDocument123 pagesلیست کد های ATCEman ShalabyNo ratings yet
- Antiinflamatoare NesteroidieneDocument2 pagesAntiinflamatoare NesteroidieneLore DanaNo ratings yet
- Materias Primas Prohibidas Por KimberlyDocument7 pagesMaterias Primas Prohibidas Por KimberlyRoberto BonillaNo ratings yet
- Density Values of Several PlasticsDocument4 pagesDensity Values of Several PlasticsAbdullah Al AsikNo ratings yet
- BIO CHEM Multiple Choice QuestionsDocument40 pagesBIO CHEM Multiple Choice QuestionsSp PpvNo ratings yet
- Selinium ReagentDocument18 pagesSelinium ReagentShiv GuptaNo ratings yet
- AMS Guide To Equivalent HRT DosesDocument3 pagesAMS Guide To Equivalent HRT DosesuioNo ratings yet
- Functional Group NamesDocument21 pagesFunctional Group NamesAdine RaissaNo ratings yet
- FYNK PharmaceuticalsDocument13 pagesFYNK PharmaceuticalsHina TinaNo ratings yet
- CBSE Class 12 Physics Important Questions Polymers: Material Downloaded From - 1 / 5Document5 pagesCBSE Class 12 Physics Important Questions Polymers: Material Downloaded From - 1 / 5Rahil ShamsiNo ratings yet
- 1 of 33 © Boardworks LTD 2009Document33 pages1 of 33 © Boardworks LTD 2009gabbbbbbbbbbbbbbbbNo ratings yet
- Daftar Nama Obat SIMPUSDocument11 pagesDaftar Nama Obat SIMPUSsdk.achmad TubeNo ratings yet
- This Study Resource Was: DNA Base Pairing WorksheetDocument5 pagesThis Study Resource Was: DNA Base Pairing Worksheetphil tolentinoNo ratings yet
- 5 - Biochemistry MCQs Cetric Acid CycleDocument9 pages5 - Biochemistry MCQs Cetric Acid CycleSantosh Bhandari100% (1)
- Brosur LabkesdaDocument1 pageBrosur Labkesdael_karimahNo ratings yet
- Light-Dependent Herbicides - An OverviewDocument12 pagesLight-Dependent Herbicides - An OverviewCatherine TangNo ratings yet