Professional Documents
Culture Documents
Origin of Replication: Gram Negative Bacterial Plasmids
Uploaded by
Magesh Ramasamy0 ratings0% found this document useful (0 votes)
122 views12 pagesPlasmids can replicate independently of the host chromosome using their own origin of replication. The two main modes of plasmid replication are theta replication and rolling circle replication. Plasmid replication, host range, copy number, and incompatibility are controlled. The ColE1 plasmid replicates using RNA primer-dependent theta replication controlled by antisense RNA regulation. RNA I binds to RNA II to interfere with its structure and ability to prime replication. The Rop protein enhances binding of RNA I to RNA II to regulate ColE1 copy number.
Original Description:
Original Title
Copy Number Control of ColE1 Plasmid
Copyright
© © All Rights Reserved
Available Formats
PPT, PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentPlasmids can replicate independently of the host chromosome using their own origin of replication. The two main modes of plasmid replication are theta replication and rolling circle replication. Plasmid replication, host range, copy number, and incompatibility are controlled. The ColE1 plasmid replicates using RNA primer-dependent theta replication controlled by antisense RNA regulation. RNA I binds to RNA II to interfere with its structure and ability to prime replication. The Rop protein enhances binding of RNA I to RNA II to regulate ColE1 copy number.
Copyright:
© All Rights Reserved
Available Formats
Download as PPT, PDF, TXT or read online from Scribd
0 ratings0% found this document useful (0 votes)
122 views12 pagesOrigin of Replication: Gram Negative Bacterial Plasmids
Uploaded by
Magesh RamasamyPlasmids can replicate independently of the host chromosome using their own origin of replication. The two main modes of plasmid replication are theta replication and rolling circle replication. Plasmid replication, host range, copy number, and incompatibility are controlled. The ColE1 plasmid replicates using RNA primer-dependent theta replication controlled by antisense RNA regulation. RNA I binds to RNA II to interfere with its structure and ability to prime replication. The Rop protein enhances binding of RNA I to RNA II to regulate ColE1 copy number.
Copyright:
© All Rights Reserved
Available Formats
Download as PPT, PDF, TXT or read online from Scribd
You are on page 1of 12
Plasmid Replication
Plasmids must be able to replicate independently of
the host chromosome. They have their own origin of
replication (ori).
There are two general modes of replication:
1) THETA Replication -- which is found in most of the
gram negative bacterial plasmids such as ColE1,
RK2, and F
2) ROLLINGCIRCLE Replication -- which is found in
some gram positive bacterial plasmids such pLS1,
pLB4
Replication is central to control number of important
plasmid properties:
HOST RANGE, COPY NUMBER, INCOMPATIBILITY and
Plasmid Host Range
Refers to plasmids ability to survive/replicate in a particu
host.
Some plasmids are able to replicate in a limited numbe
bacterial species; they have a NARROW host range.
Examples: ColE1, pBR322, pUC18 plasmids which are lim
to E.coli and some closely related species.
Other plasmids are able to replicate in a wide range of
bacterial species; they have a BROAD host range.
Eg: RSF1010 and RK2
Copy number control of
ColE1 Plasmid
The number of copies of a plasmid can vary from 1 ( F
plasmid) to 100 (pUC18).
Copy number is an important property of the plasmid
and depends on the mechanism by which it regulates
its own replication.
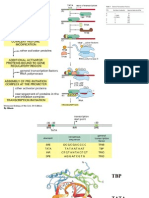
Replication control of ColE1 Plasmid
ColE1 is a small circular plasmid which codes for a 57
kDal protein toxin (colicin E1) which can kill other
E.coli cells.
It carries (oriV) origin of replication and a region
(bom) at which it can be mobilized for transfer to
other bacteria.
Other Genes of ColE1 Plasmid and their
functions as follows:
Circular Map of ColE1 Plasmid
Replication of ColE1 proceeds unidirectionally from
oriV and depends on RNA II which is transcribed from a
promoter 555 bp away from oriV.
When this transcript reaches oriV, one of two possible
fates is possible:
The RNA-DNA hybrid is cleaved by RNaseH to
generate a free 3'-OH end which serves as a primer for
DNA polymerase I to initiate replication.
Then DNA polymerase III takes over after a short
distance.
If RNA-DNA hybrid is NOT cleaved by RNaseH and
transcription continues nonproductively.
These fates depends on the formation of a specific
secondary structure in the nascent RNA II transcript;
If the structure forms correctly then RNaseH
processing occurs correctly.
If anything interfere with the formation of correct
secondary structure will interfere with replication
A specific RNA called RNA I whose function is to
interfere with the secondary structure of RNA II
This RNA I is a 108 nt molecule which is transcribed in the
opposite direction to RNA II from a promoter located
between the RNA II promoter and the origin of replication
Since RNA I is complementary to RNA II, it form a
stable RNA-RNA hybrid with it. If it does, then RNA II
will not be able to function as a primer.
This is an example of antisense RNA
regulation.
Replication of ColE1 also depends on the Rop
protein
Rop is a small protein (63 aas) which increases the
rate of binding of RNA I to RNA II. If Rop is absent,
plasmid replication will be more frequent.
You might also like
- VectorsDocument73 pagesVectorsZohra SafdariNo ratings yet
- Plasmids ReplicationDocument6 pagesPlasmids ReplicationRaquel VieiraNo ratings yet
- Bacteriophages: - A Class of Viruses, Which Infect Specific Bacterial CellsDocument65 pagesBacteriophages: - A Class of Viruses, Which Infect Specific Bacterial Cellsubuntu 13.04No ratings yet
- EMMRC Lambda Phage 1Document18 pagesEMMRC Lambda Phage 1Polu ChattopadhyayNo ratings yet
- SYBT TranscriptionDocument69 pagesSYBT TranscriptionMeir SabooNo ratings yet
- Structure of RetrovirusDocument12 pagesStructure of RetrovirusMatin Ahmad KhanNo ratings yet
- TranscriptionDocument10 pagesTranscriptionHardik ManekNo ratings yet
- Molecular Biology Lecture 11Document38 pagesMolecular Biology Lecture 11Anton IrvingNo ratings yet
- Virus Transcription, Translation and Transport: ST NDDocument5 pagesVirus Transcription, Translation and Transport: ST NDLana AhmedNo ratings yet
- TranscriptionDocument21 pagesTranscriptionYehya JaberNo ratings yet
- Prokaryotic TranscriptionDocument14 pagesProkaryotic TranscriptionAjay RawatNo ratings yet
- LECT-09 Transcription in EukaryotesDocument27 pagesLECT-09 Transcription in EukaryotesFAKHRI HUSAINI NASUTION 1No ratings yet
- Bacteriophage LambdaDocument45 pagesBacteriophage Lambdasourov546vermaNo ratings yet
- Lambda PhageDocument21 pagesLambda PhageMagesh RamasamyNo ratings yet
- Lec 7 TranscriptionDocument30 pagesLec 7 TranscriptionYasmin BalochNo ratings yet
- Transcription and RNA Processing in EukaryotesDocument13 pagesTranscription and RNA Processing in EukaryotesAbdulfattah NoorNo ratings yet
- Transcription in Prokaryotes: Single RNA PolymeraseDocument29 pagesTranscription in Prokaryotes: Single RNA PolymeraseShubhamNo ratings yet
- DNA TranscriptionDocument25 pagesDNA TranscriptionAkss ShauryaNo ratings yet
- Transcription in Prokaryotes and Eukaryotes: (Frequently Asked Questions)Document6 pagesTranscription in Prokaryotes and Eukaryotes: (Frequently Asked Questions)Asif AhmedNo ratings yet
- Lecture8 DNA-Dependent RNA Synthesis-2Document50 pagesLecture8 DNA-Dependent RNA Synthesis-2Constance WongNo ratings yet
- Eukaryotic TranscriptionDocument14 pagesEukaryotic TranscriptionRaj Singh dhavanNo ratings yet
- Transcription, Translation and TransportDocument16 pagesTranscription, Translation and Transportfaria11-6565No ratings yet
- Meredith Barb Homework-2 (25 Points) Due On Sept. 24 by 5:00pmDocument5 pagesMeredith Barb Homework-2 (25 Points) Due On Sept. 24 by 5:00pmMeredith BarbNo ratings yet
- E.coli RNA PolymeraseDocument17 pagesE.coli RNA PolymeraseM.PRASAD NAIDUNo ratings yet
- Messenger Rnas (Mrnas) : This Class of Rnas Are The Genetic Coding TemplatesDocument8 pagesMessenger Rnas (Mrnas) : This Class of Rnas Are The Genetic Coding TemplatesRavi AlugubelliNo ratings yet
- Assignment On Regulation of Transcription, Lytic Lysogeny Cascade and SOS Regulatory SystemDocument16 pagesAssignment On Regulation of Transcription, Lytic Lysogeny Cascade and SOS Regulatory SystemShraddha Bhatt ChavanNo ratings yet
- 11 Chapter 5Document38 pages11 Chapter 5toobashafiNo ratings yet
- 4 Gene ExpressionDocument35 pages4 Gene ExpressionThảo ThảoNo ratings yet
- Lambda PhageDocument4 pagesLambda PhagesauravNo ratings yet
- BMM GM2 ADocument6 pagesBMM GM2 ASleepyHead ˋωˊNo ratings yet
- BIOCORE 1 Midterm 2 Study GuideDocument44 pagesBIOCORE 1 Midterm 2 Study GuideWarren EnglishNo ratings yet
- Transcription EukarDocument3 pagesTranscription EukarnitralekhaNo ratings yet
- Dna L12 NotesDocument6 pagesDna L12 NotesellieNo ratings yet
- II TranscriptionDocument46 pagesII TranscriptionGail IbanezNo ratings yet
- TranscriptionDocument56 pagesTranscriptionVipin100% (8)
- TranscriptionDocument61 pagesTranscriptiondeepak mauryaNo ratings yet
- LCV Q2 Lecture 1Document58 pagesLCV Q2 Lecture 1Abigail SaballeNo ratings yet
- Molecular Biology NotesDocument8 pagesMolecular Biology Notesbyunus88No ratings yet
- Genetics Exam ReviewerDocument9 pagesGenetics Exam ReviewerEysi BarangganNo ratings yet
- TransformationDocument24 pagesTransformationSai SridharNo ratings yet
- Molecular Biology For Forth SemDocument10 pagesMolecular Biology For Forth Semmahammad rizwanNo ratings yet
- Biology NotesDocument4 pagesBiology Noteselena m.No ratings yet
- TranscriptionDocument23 pagesTranscriptionareen fakhouryNo ratings yet
- RNA Synthesis, Processing SBT 211Document59 pagesRNA Synthesis, Processing SBT 211TADIWANASHE TINONETSANANo ratings yet
- L8. Transcription TranslationDocument15 pagesL8. Transcription TranslationDiane Princess SultanNo ratings yet
- Tema 7Document11 pagesTema 7Lucía Alba OrdoñezNo ratings yet
- TranscriptionDocument20 pagesTranscriptionlordniklausNo ratings yet
- Molecular Cell Biology Lodish 6th Edition Test Bank DownloadDocument9 pagesMolecular Cell Biology Lodish 6th Edition Test Bank Downloadjohndanielswrydmzokge100% (26)
- Transcription ProkaryoticDocument30 pagesTranscription ProkaryoticDibya Jyoti ParidaNo ratings yet
- Tarnscription RNA POLYMERASEDocument26 pagesTarnscription RNA POLYMERASEnarasimmareddy.cNo ratings yet
- Transcription by RNA Polymerases I and IIIDocument16 pagesTranscription by RNA Polymerases I and IIIxilax99No ratings yet
- What Is RNA PolymeraseDocument15 pagesWhat Is RNA Polymerasemohmedmustafa4No ratings yet
- BOCM ST2 TUT QuestionsDocument4 pagesBOCM ST2 TUT QuestionsntsakopreciouskoNo ratings yet
- DNA Transcription (Part-1) : DR - Ahmed Salim Mohammed PH.D Molecular MicrobiologyDocument49 pagesDNA Transcription (Part-1) : DR - Ahmed Salim Mohammed PH.D Molecular MicrobiologyDrAhmedSalimNo ratings yet
- Principles of Microbiology 1 (Summary)Document8 pagesPrinciples of Microbiology 1 (Summary)Mabelle DucusinNo ratings yet
- Transcriprion ManuscriptDocument14 pagesTranscriprion ManuscriptAsif AhmedNo ratings yet
- Reverse TranscriptionDocument24 pagesReverse TranscriptionGela CandelariaNo ratings yet
- The Molecular Biology of Plastids: Cell Culture and Somatic Cell Genetics of PlantsFrom EverandThe Molecular Biology of Plastids: Cell Culture and Somatic Cell Genetics of PlantsIndra VasilNo ratings yet
- BPT Chromatography TechniquesDocument48 pagesBPT Chromatography TechniquesMagesh RamasamyNo ratings yet
- PlasmidsDocument14 pagesPlasmidsMagesh RamasamyNo ratings yet
- Lambda PhageDocument21 pagesLambda PhageMagesh RamasamyNo ratings yet
- MPN TableDocument1 pageMPN TableMagesh RamasamyNo ratings yet
- TIMEMANAGEMENTDocument13 pagesTIMEMANAGEMENTPolinda UseroNo ratings yet
- Formation and Destruction of Red Blood Cells, and The Recycling of Hemoglobin ComponentsDocument1 pageFormation and Destruction of Red Blood Cells, and The Recycling of Hemoglobin ComponentsK-Alert R-Man100% (2)
- Vaishnavi Sing: Zulekha Hospital LLC - (SHARJAH)Document1 pageVaishnavi Sing: Zulekha Hospital LLC - (SHARJAH)Abc AbcNo ratings yet
- The Brain Adapts To Dishonesty: Neil Garrett, Stephanie C Lazzaro, Dan Ariely & Tali SharotDocument9 pagesThe Brain Adapts To Dishonesty: Neil Garrett, Stephanie C Lazzaro, Dan Ariely & Tali SharotVissente TapiaNo ratings yet
- Woody Plant Seed ManualDocument1,241 pagesWoody Plant Seed ManualElena CMNo ratings yet
- NUTR4320FINAL11Document10 pagesNUTR4320FINAL11rijzNo ratings yet
- Neuro Informo1Document8 pagesNeuro Informo1Dr. Kaushal Kishor SharmaNo ratings yet
- Olfactory Colours and Coloured SmellsDocument28 pagesOlfactory Colours and Coloured Smellsspa100% (2)
- JGXP - 2011 - v15n4 - Contamination Control in The Compliance Program PDFDocument7 pagesJGXP - 2011 - v15n4 - Contamination Control in The Compliance Program PDFNelson Alejandro FierroNo ratings yet
- Potometer Set UpDocument2 pagesPotometer Set UpEliseo PamandananNo ratings yet
- S. 3 Biology Paper 1Document9 pagesS. 3 Biology Paper 1Nsaiga RonaldNo ratings yet
- AQUATICS Keeping Ponds and Aquaria Without Harmful Invasive PlantsDocument9 pagesAQUATICS Keeping Ponds and Aquaria Without Harmful Invasive PlantsJoelle GerardNo ratings yet
- Iceberg ModelDocument1 pageIceberg ModelMaria Melody GadoNo ratings yet
- Short Questions Guess Paper For FSC Part-1 Biology: 1st Year Biology Guess 2018 (All Punjab Boards) Unit 1Document4 pagesShort Questions Guess Paper For FSC Part-1 Biology: 1st Year Biology Guess 2018 (All Punjab Boards) Unit 1Umer Asfandyar BalghariNo ratings yet
- Alfa Laval Disc Stack Centrifuge TechonologyDocument8 pagesAlfa Laval Disc Stack Centrifuge TechonologyChaitanya B.AndhareNo ratings yet
- Evs ProjectDocument3 pagesEvs ProjectPallavi BanerjeeNo ratings yet
- Use of Dinitrosalicylic Acid Reagent For Determination of Reducing SugarDocument7 pagesUse of Dinitrosalicylic Acid Reagent For Determination of Reducing SugarLANANo ratings yet
- Fitness Leader's HandbookDocument623 pagesFitness Leader's HandbookRaviNo ratings yet
- Hamstring TendinopathyDocument16 pagesHamstring TendinopathySaravananNo ratings yet
- Presentation1 1Document15 pagesPresentation1 1Manula MuthunayakeNo ratings yet
- Adult Development and AgingDocument44 pagesAdult Development and AgingDiana DayenNo ratings yet
- Detailed Lesson Plan in ScienceDocument8 pagesDetailed Lesson Plan in ScienceRegine MalanaNo ratings yet
- 12 Biology Notes Ch11 Biotechnology Principles and ProcessesDocument9 pages12 Biology Notes Ch11 Biotechnology Principles and ProcessesAnkit YadavNo ratings yet
- Mechanisms of Perception Hearing, Touch, Smell, Taste and AttentionDocument22 pagesMechanisms of Perception Hearing, Touch, Smell, Taste and Attentiontomiwa iluromiNo ratings yet
- Banana Expert System - Crop ProtectionDocument38 pagesBanana Expert System - Crop ProtectionbhushanNo ratings yet
- Catalogo Nuevo GboDocument7 pagesCatalogo Nuevo GboCaro ErazoNo ratings yet
- Higuchi-1961-Journal of Pharmaceutical Sciences PDFDocument2 pagesHiguchi-1961-Journal of Pharmaceutical Sciences PDFEllya AgustinaNo ratings yet
- Mareschal Et Alii, 2007, Chapter 5 Sirois Et Alii, 2008, P. 325, and Figure 1 Below, Section IIIDocument15 pagesMareschal Et Alii, 2007, Chapter 5 Sirois Et Alii, 2008, P. 325, and Figure 1 Below, Section IIIHenrique Augusto Torres SimplícioNo ratings yet
- Pengaruh Pemberian Infusa Daun KemuningDocument66 pagesPengaruh Pemberian Infusa Daun Kemuningvidianka rembulanNo ratings yet
- Common Model Exam Set-XV (B) (2079-4-14) QuestionDocument16 pagesCommon Model Exam Set-XV (B) (2079-4-14) QuestionSameer KhanNo ratings yet